A shorter, sharper Out-of-Africa story is emerging
A new study of genomic ancestry in Papua New Guinea supports a short timeline for the dispersal of modern people
Over the last few months I’ve been working to understand a potentially epochal finding from earlier this year. Genetic data are suggesting a “hard ceiling” on the time of dispersal of modern people across Eurasia. If the models are correct, the dispersals proceeded very fast but only after around 47,000 years ago.
But there’s a problem: A dispersal after 47,000 years ago seems to contradict some archaeological and skeletal evidence. Archaeologists have been pushing the timeline of modern human dispersal into East and island Southeast Asia further and further back. Some argue that Australia was occupied before 65,000 years ago, pictorial art on Sulawesi dates to 51,000 years ago or older, and some sites on mainland East and Southeast Asia have skeletal or dental remains that look modern but are older than 60,000 years—and in a couple of cases, more than 100,000.
If such evidence doesn’t represent early modern humans, who were these people?
A new paper recently came across my desk that narrows the possible solutions. Mayukh Mondal and collaborators in their new work focused on the DNA of people from Papua New Guinea. The researchers used whole genome data to test when Papuan people shared common ancestors with the populations of mainland East Asia, finding that time to be an estimated 46,200 years ago.
That time may still be a bit early for a dispersal that may only have started after 47,000 years ago. Still, it’s in the ballpark. What I find significant is that this estimate comes from a different line of analysis—one that a few years ago seemed to suggest a much older timeline. The new results sink an alternative explanation that was floated almost a decade ago: a “ghost modern” population in Southeast Asia.
Disposing of a ghost population
Back in 2016, Luca Pagani and coworkers studied whole-genome data drawn from a sample of worldwide populations, including some individuals from Papua New Guinea. Theirs was an early example of an analysis using a statistic called the cross-coalescence rate, which looks at when population-specific haplotypes in genomes from two populations are estimated to have come from common ancestral population. Cross-coalescence has become a valuable tool in reconstructing the divergence of populations. From the data they could estimate split times between present-day populations represented by genome data.
Pagani and collaborators compared the Papuan genomes to those from the Yoruba population of present-day Nigeria—one of the best-represented African populations in genome datasets. They estimated that these populations began to separate around 90,000 years ago.
That’s long before 50,000 years ago.
More interesting, this estimate for PNG was substantially earlier than Pagani and coworkers estimated for Yoruba and mainland Eurasian populations, which seemed to have split around 75,000 years ago.
What could explain Papuan people getting an earlier start from Africa than any mainland groups?
One possibility was that the older split time for PNG actually might reflect their higher Denisovan ancestry. Denisovans really did diverge from African groups a very long time ago, so a small fraction of Denisovan DNA ancestry might make Papuan genomes look like they separated from Africans well before the actual time of the massive dispersal across Eurasia. Pagani and coworkers tested for this possibility by “masking” the portions of each genome that were most similar to the Denisova 3 genome. The effect remained even without this Denisovan mixture.
This left another possibility: Maybe another ghost population had contributed to the PNG genomes. This ghost was modern, much closer to both African and Eurasian genomes than Denisova 3. But it had separated from African groups long before the split of African from today’s Eurasian populations: maybe around 150,000 years ago. As Pagani and coworkers described it, this was an early extinct out-of-Africa (xOoA) population had existed in Asia.
“We find a genetic signature in present-day Papuans that suggests that at least 2% of their genome originates from an early and largely extinct expansion of anatomically modern humans (AMHs) out of Africa.”—Luca Pagani and coworkers
That idea was appealing to many anthropologists. Skeletal finds from China seemed to show that people with “modern” anatomical features had been present in the region as early as 100,000 years ago or more. Such a population would have existed long before the mixture with Neanderthals and Denisovans, so clearly a much later dispersal would have been responsible for most of the heritage of today’s Asian peoples. But if there was a “ghost” population in the genomes, maybe such early fossils were traces of it.
Yet there was immediately skepticism from many geneticists. This one approach using cross-coalescence models made it seem like a ghost population was a possible solution, but the genomes showed no other obvious signs of extinct out-of-Africa mixture. Researchers were getting better and better at finding small traces of introgressed genome, and they just didn’t see it.
An exception was a 2022 research article by Anthony Wohns and collaborators, who tried to build a giant family tree connecting more than 3000 human genomes. Their innovative method sketched out the possible geographic locations of common ancestors within that family tree. A handful of those common ancestors were inferred to have been present in Papua New Guinea by an estimated 140,000 years ago. The authors noted that those ancestors could have been part of an extinct out-of-Africa ghost population. On the other hand, they saw, maybe deeply-diverged Denisovan ancestors could explain this part of their results.
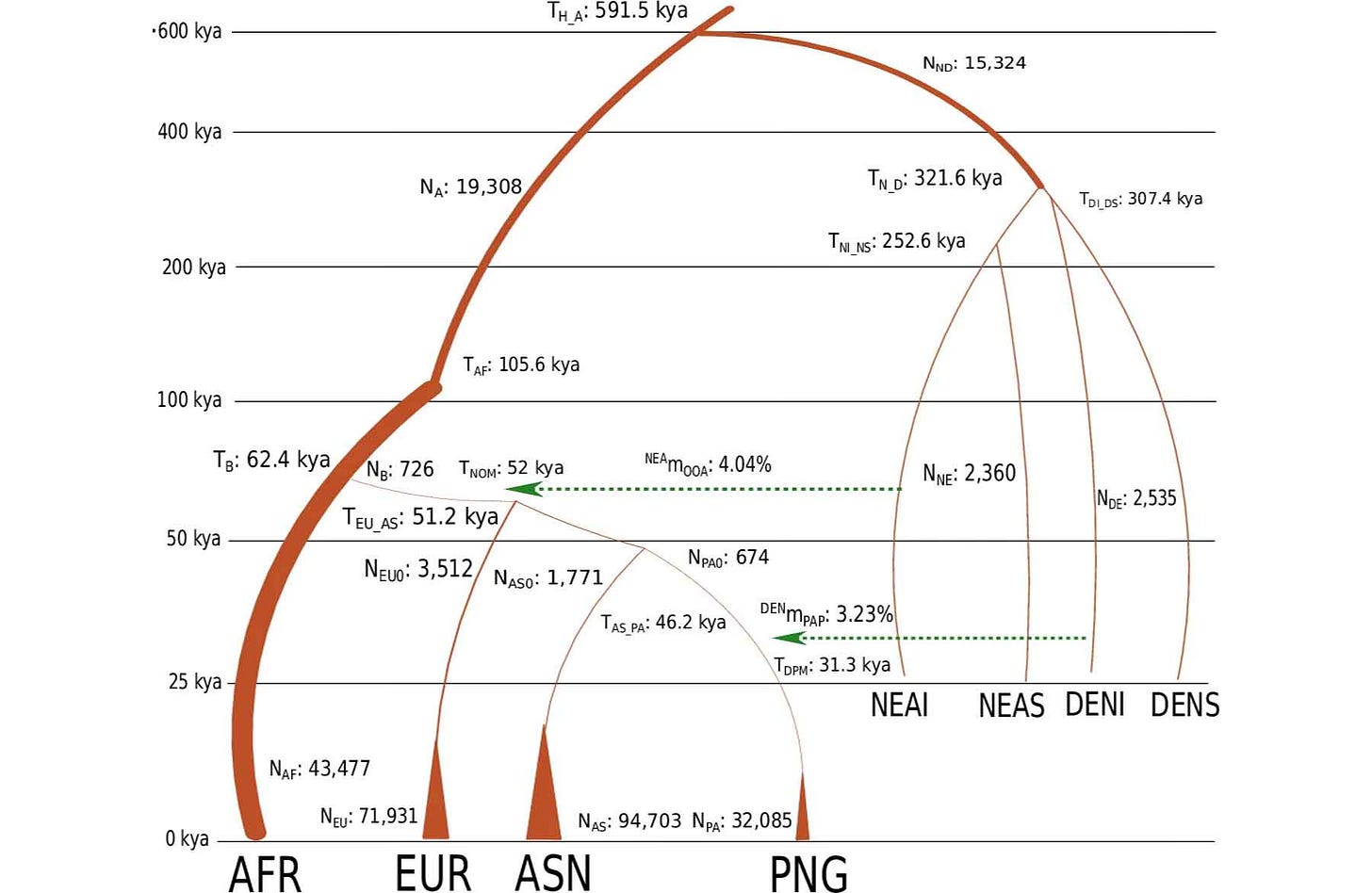
The new work by Mondal and collaborators establishes a better explanation for the cross-coalescence results. By modeling a strong bottleneck at the founding of the Papuan population, the new study was able to replicate the relative cross-coalescence results of Pagani and coworkers. In a nutshell, it wasn’t a contribution from an early ghost, it was a bottleneck followed by slow population growth that inflated the appearance of a population separation.
This is a persuasive explanation for the previous findings. None of these results pointing to a ghost population were ever about the raw data; every conclusion came from certain decisions within the modeling of ancient population sizes and times of divergence. These papers share a number of authors, and the cross-checking of results was one of the main aims of the new paper.
Meeting Denisovans
The new research by Mondal and collaborators also shows that mixture of Denisovans into the ancestry of Papuan groups came—at least in part—a lot later than the separation of ancestors of today’s Papuan and East Asian populations. The study estimates 31,000 years ago for Denisovan mixture into the Papuan ancestral population.
This isn’t the first estimate of this kind of recent Denisovan mixture. The most comprehensive result came from a study by Guy Jacobs and collaborators in 2019. Jacobs and coworkers studied both people from Papua New Guinea and from the Indonesian portion of the island, West Papua. They showed that there had been three or more distinct episodes of Denisovan mixture within the histories of varied populations. The diversity of mixture from these different Denisovan populations is greatest in some of today’s groups from PNG and others from the Philippines. The study showed that two distinct groups were represented in the Papuan genomes, which Jacobs and coworkers named D1 and D2.
The D1 mixture was estimated to have happened around 30,000 years ago, with a wide confidence interval from 14,000 to 50,000. The D2 mixture was more widespread, present in a wider array of populations in island Southeast Asia. It was also around 50% earlier, an estimated 46,000 years ago—again with a wide confidence interval stretching from 32,000 to 60,000 years ago.
The new work by Mondal and coworkers does not probe the more complex scenario presented by Jacobs and coworkers back in 2019. From the point of view of cross-coalescence analysis of modern human groups, the important thing is to recognize Denisovan and Neanderthal haplotypes within the genome and mask them off—the deeply diverged nature of these introgressed haplotypes would otherwise tend to inflate estimates of population divergence. In my view, the older results by Jacobs and team still provide the most accurate idea of what Denisovan ancestry was like in Papuan groups. The new results help to reinforce that recent Denisovan mixture really did take place in this region.
Together, these findings suggest strongly that one or more Denisovan groups were already present in Wallacea or even Sahul when modern humans entered the region. That, too, may help to explain some of the apparent archaeological evidence for early human entry into Papua New Guinea, such as Mololo Cave or Australian sites like Madjedbebe: Denisovans may have been there first.
What about those fossils?
One thing I want to add at the bottom of this post: All this discussion about the genomic ancestry of Papuan groups, their split times from mainland Asian populations, and the Denisovans adds nothing new to the problem of the fossil record of mainland East and Southeast Asia. Those modern-looking skeletal remains are still there.
I have to acknowledge that there has been some strong debate about the geochronology of some of the key sites, like Fuyan Cave, or the morphology of the fossils, such as Zhiren Cave. Specialists in geochronological dating of rock art have rarely come to any consensus about early dates from any site. Still, there are a lot of these finds, and some of them—including the early modern human finds from Tam Pà Ling, Laos—are very solid. Yet these appear far too ancient to derive from a founding population only 47,000 years ago or less.
There may really have been an extinct out-of-Africa population in this region. They could have been connected with groups that are represented in southwest Asia at Skhūl and Qafzeh Caves, for example.
On the other hand, maybe some Denisovan groups were much more modern-looking than most researchers may be assuming. It would be a simple explanation of the genomic findings, after all. Groups with primarily Denisovan heritage, maybe with multiregional gene flow from contemporaries in Africa or Southwest Asia, may have been diverse in body size, cranial form, and behavior. The behavioral sophistication of later Neanderthals has been clear now for decades. Cultural innovations by later Denisovans—who lived in a broader array of ecologies—may have been even more profound.
References
Brucato, N., André, M., Tsang, R., Saag, L., Kariwiga, J., Sesuki, K., Beni, T., Pomat, W., Muke, J., Meyer, V., Boland, A., Deleuze, J.-F., Sudoyo, H., Mondal, M., Pagani, L., Gallego Romero, I., Metspalu, M., Cox, M. P., Leavesley, M., & Ricaut, F.-X. (2021). Papua New Guinean Genomes Reveal the Complex Settlement of North Sahul. Molecular Biology and Evolution, 38(11), 5107–5121. https://doi.org/10.1093/molbev/msab238
Mondal, M., André, M., Pathak, A. K., Brucato, N., Ricaut, F.-X., Metspalu, M., & Eriksson, A. (2025). Resolving out of Africa event for Papua New Guinean population using neural network. Nature Communications, 16(1), 6345. https://doi.org/10.1038/s41467-025-61661-w
Sümer, A. P., Rougier, H., Villalba-Mouco, V., Huang, Y., Iasi, L. N. M., Essel, E., Mesa, A. B., Furtwaengler, A., Peyrégne, S., de Filippo, C., Rohrlach, A. B., Pierini, F., Mafessoni, F., Fewlass, H., Zavala, E. I., Mylopotamitaki, D., Bianco, R. A., Schmidt, A., Zorn, J., … Krause, J. (2024). Earliest modern human genomes constrain timing of Neanderthal admixture. Nature, 1–3. https://doi.org/10.1038/s41586-024-08420-x
Pagani, L., Lawson, D. J., Jagoda, E., Mörseburg, A., Eriksson, A., Mitt, M., Clemente, F., Hudjashov, G., DeGiorgio, M., Saag, L., Wall, J. D., Cardona, A., Mägi, R., Sayres, M. A. W., Kaewert, S., Inchley, C., Scheib, C. L., Järve, M., Karmin, M., … Metspalu, M. (2016). Genomic analyses inform on migration events during the peopling of Eurasia. Nature, 538(7624), 238–242. https://doi.org/10.1038/nature19792
Wohns, A. W., Wong, Y., Jeffery, B., Akbari, A., Mallick, S., Pinhasi, R., Patterson, N., Reich, D., Kelleher, J., & McVean, G. (2022). A unified genealogy of modern and ancient genomes. Science, 375(6583), eabi8264. https://doi.org/10.1126/science.abi8264


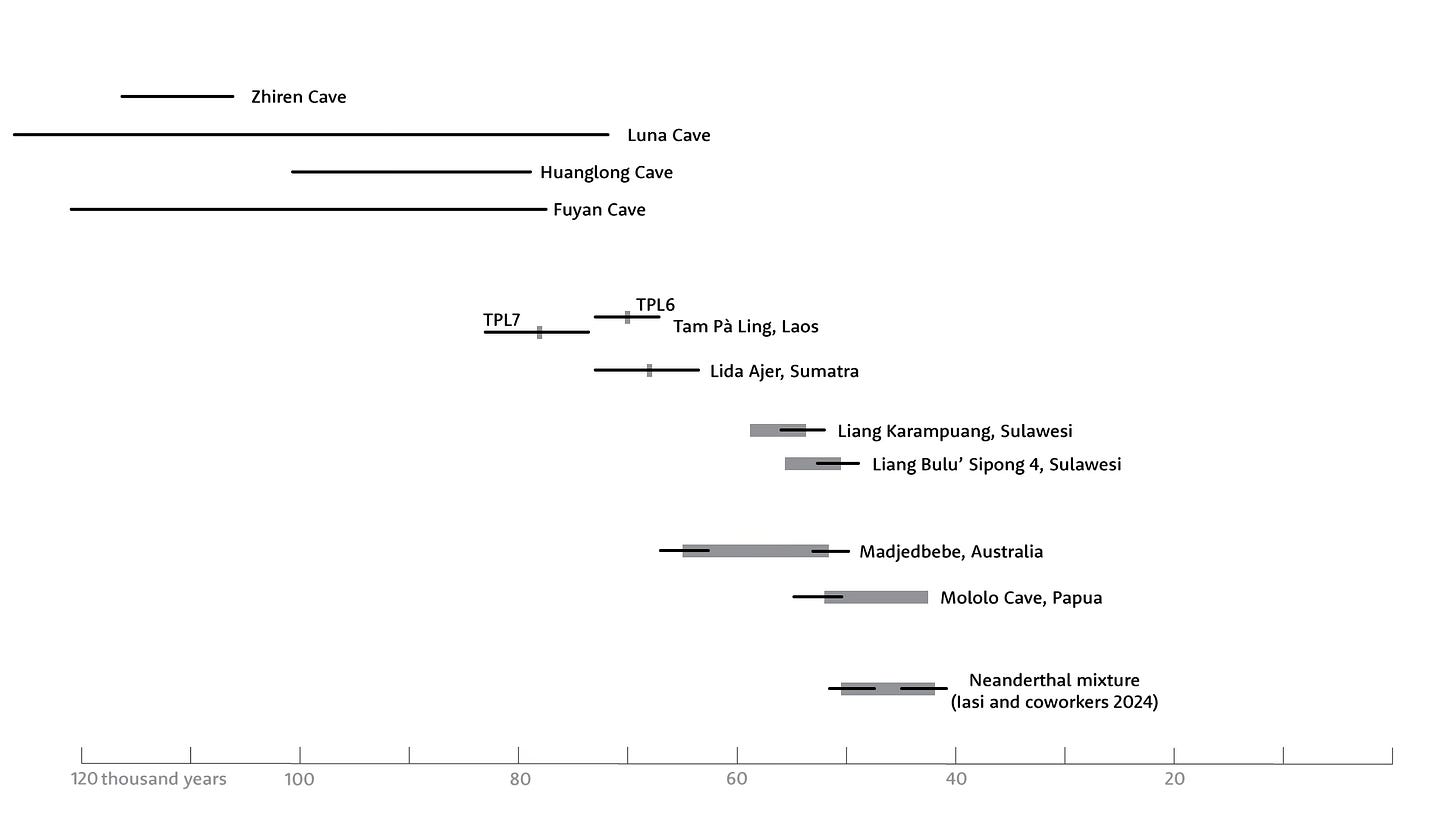
This post is such a good summary of the "wallasea problem".
Yet I am not convinced by the Mondal et al work.
First, even by stretching their model parameters to the max, they obtained only a small difference in coalescence times between PNG-Afr and Asi-Afr compared to real data, (compare fig 1 and 4 from their paper).
Second, surely there must be other Eurasian populations that could experience strong bottlenecks and long isolation. Yet the coalescence time between all Eurasians and Afr Yoruba is very consistent, and doesn't exceeds 75 my, that is likely happened after Toba.
I thing the right explanation is the most obvious one -that there was an early HS population in SEA that was mostly but not fully replaced by newcomers after 50 ky but left some genetic trace.
John, is my interpretation of the RCCR method correct here?
Say you have two populations, popA and popB, with an RCCR value of 0.5 (on a scale from 0 to 1) at 50k ybp. Does that essentially mean that 50% of the unique drift that defines popA as popA distinct from popB (and vice versa) had accumulated by 50k ybp?
Looking at "Table 1 Best fitted parameter values of model A," they have the separation of African and OOA populations at 62k ybp, with a 10k year long bottleneck. The estimated effective population size of OOA during that time is only 726 - that seems like a very, very low population size to be able to ride out a 10k year bottleneck without just going extinct.