Ghost populations in human origins
Genetic models are finding more and more unknown lineages. How real are they?

Every person’s genome has a few stretches of DNA that stand out as different from most of humanity. Some are traces of ghost populations: ancient groups that became extinct, but not before contributing some of their own genes to other populations that survived.
Geneticists have identified some of these ghosts. Everyone shares some of these quirky sequences with Neanderthals or Denisovans, long-lost groups that persist today only within living people’s genomes. But other groups remain a mystery.
Ideas about ghost populations have become central to our understanding of human origins and evolution of other mammals. I’ve written about ghost hominins, ghost bonobos, and ghost gorillas.
Even so, the idea covers up a lot of uncertainty about how human groups were connected with each other in the past. Every “ghost” is a mathematical construct waiting for DNA from ancient skeletons to test whether it was real.
So let’s look at the current reality of these genetic ghost ancestors.
Ghost hunting history
Ghost populations are deduced from statistics, with data from living people and from ancient skeletons where it exists. When two populations divide from each other for a long time and then come back together, telltale signs mark the genomes of their descendants, even after thousands of years.
Mutations that are near each other on the same chromosome tend to be inherited together. This genetic linkage results in barcode-like arrangements of mutations across thousands of base pairs known as haplotypes. If a population never changed in size and never interacted with distant relatives, then haplotype lengths, haplotype frequencies, and mutational diversity all would tend toward a predictable pattern.
But when a very different population enters the mix, it has its own unique set of haplotypes that have not remixed with those in the first population. They’re long, they have different mutations, and that makes them stick out.
Geneticists started noticing such long, divergent haplotypes in living human populations during the 1990s. When I first entered human genetics research, I wondered whether some of these divergent haplotypes came from Neanderthals. I was far from alone. Several research groups in the early 2000s were taking a close look at genes with long, divergent haplotypes.

One of my collaborative projects focused on a gene called MCPH, which had a long haplotype that didn't seem to fit with the idea that humans had come from a single small ancestral group. We guessed it was likely Neanderthal in origin.
Another research team led by Michael Hammer focused on two regions of the X chromosome that likewise showed very deep haplotype diversity, both in East Asian populations. That was a bit puzzling. The possible source wasn’t clear at the time. No one used the term “ghost population” back then, but today we would call this a sign of one.
Meanwhile, a few groups of geneticists were beginning to develop statistics to handle thousands of different haplotypes across the whole genome. I remember being blown away by a groundbreaking paper in 2006 written by Vincent Plagnol and Jeffrey Wall. They estimated that 5% of European ancestry came from some kind of ghost population. To scientists like me, that conclusion made good sense: this ghost population must have been the Neanderthals.
What was harder to explain was Plagnol and Wall’s result looking at the genomes of West African people. They, too, had inherited around 5% of their genomes from some divergent ancestral group. Neanderthals didn’t seem likely—no one had ever found them in Africa. This was the first strong evidence pointing to a ghost population in human ancestry.
“[A]rchaic populations such as Neanderthals must have made a substantial contribution to the modern gene pool in Europe. We observe a similar pattern for West African populations even though a clear source population has not yet been found.”—Vincent Plagnol and Jeffrey Wall
Pinning down ghosts with ancient DNA
In those early days, skeptical geneticists were hard to convince about such ancestors. Then in 2010 the Neanderthal genome appeared. Instantly it became clear that most people with heritage from Europe, Asia, the Americas, and Oceania had some Neanderthal ancestors, amounting to between 1–4% of their genomes. For the divergent haplotypes, this was the equivalent of the Scooby Gang pulling off the ghost's mask and revealing a real fossil underneath.
Ancient DNA findings from Denisova Cave, also published in 2010, changed the game even further. In the lead-up to that discovery, some geneticists studying people’s genomes from Papua New Guinea had noticed what seemed like traces of mixture from an unknown group. Again they wondered, could it be Neanderthals?
The genome of Denisova 3 revealed the true answer: Denisova-like populations had once been widespread across East Asia and Southeast Asia.
In their studies of ancient and modern genomes, a team of geneticists led by David Reich and Nick Patterson innovated new kinds of analyses that sometimes pointed to ghost populations: tests of population relationships with f-statistics. These tests are especially useful for ancient genomes because they work by simply counting shared genetic variations, not relying on the lengths of haplotypes that are broken apart in fragmented DNA from ancient bones. The most informative is the f4–statistic, which considers four individuals from four populations and assesses whether their pattern of shared genetic variations fits the tree connecting the four. Another test pioneered by the same team, called the D-statistic, relies on similar logic and was also widely used to examine mixture in past groups.
When these tests show unexpected patterns of shared variation, one explanation is that one of the genomes has some ancestors from yet another group. Often the source of this ancestry can be found by applying the test again and again to more groups.
But sometimes that search comes up empty. Then there may be a ghost in the genome.
The “superarchaic” ghost
Neanderthals and Denisovans, sometimes known as archaic humans, are not ghosts. We can tie their genetic signatures to their physical remains—the oldest human remains for which this is possible today. But we have much older ancestors. There are signs that such older ancestors may have persisted and mixed with the Denisovans and Neanderthals. Geneticists have started to call such distant ghosts the superarchaics.
Superarchaics first manifested in 2014. After their first DNA discovery from the Denisova 3 finger bone, Svante Pääbo and his lab poured resources into sequencing more bone fragments from this exceptional site. One of these, a toe bone designated as Denisova 5, came from an early Neanderthal individual. To avoid confusion with the Denisova 3 “Denisovan” genome, they gave this toe a nickname: the “Altai Neanderthal”.
This Altai Neanderthal was no ghost. But it revealed one hiding in the genome from Denisova 3.
With such high-quality data from Neanderthal and Denisovan genomes for the first time, the team applied the D-statistic approach to these ancient individuals. What they found was a surprise: Denisova 3 seemed to have a small fraction of input from a vastly-more-ancient lineage.
These results came amid a burst of studies revealing repeated and widespread mixing between Neanderthals, Denisovans, and more recent people. As such work rolled out, researchers showed that Neanderthal evolution had been influenced by as much as 6–10% mixture from African sources. That Neanderthal-African mixing explained some of the D-statistic results from Denisova 3, changing what at first looked like a major superarchaic apparition into a smaller shadow.

This wasn’t the end of the superarchaic story. The anthropological geneticist Alan Rogers led a study in 2020 that gave a different spin to this ghost population. Rogers and his collaborators found that a population had divided from the mainstream of later humanity between 2.2 and 1.8 million years ago. Much later, that superarchaic group mixed at least twice into other groups: first, into the common ancestral stem that led to Neanderthal and Denisovan populations, and again later into the lineage ancestral to the Denisova 3 individual.
Who was this superarchaic ghost? One possible scenario is that this population was Homo erectus, which first entered Eurasia sometime before 1.8 million years ago. That’s my best guess.
But it’s not likely this idea will be tested anytime soon. The remains of H. erectus in Asia are tough situation for ancient DNA preservation: either much older or much warmer than scientists have recovered DNA from fossil humans before. The superarchaic may remain ghostly for a long time.
Ghost moderns and ghost archaics
Many scientists have followed in the footsteps of Plagnol and Wall looking for ancient ghosts in Africa. In 2012, Joseph Lachance and collaborators found statistical evidence of gene flow from an unknown ancestral group within the present-day Hadza and Sandawe people of Tanzania, and Baka, Bakola, and Bedzan of Cameroon. Work by Michael Hammer’s research group led by PingHsun Hsieh a few years later found a similar result from Baka and Biaka hunter-gatherer individuals.
Arun Durvasula and Sriram Sankararaman added a significant detail when they followed up with a new approach applied to the genomes of Yoruba people. Some of the diversity represented in these genomes was older than the time that Neanderthal and Denisovan populations first separated from their contemporaries in Africa.
This “ghost archaic” may not have been superarchaic, but it brought the scope of African deep mixture beyond that represented by Neanderthal or Denisovan ancestry.
“Our analyses…indicate that these populations derive 2 to 19% of their genetic ancestry from an archaic population that diverged before the split of Neanderthals and modern humans.”—Arun Durvasula and Sriram Sankararaman
Ancient DNA has begun to enrich this picture of African deep ancestry. The Shum Laka rockshelter in Cameroon contained the graves of ancient people from across the last 10,000 years, excavated starting in 1978 through the 1990s. Ten thousand years is a short time in the scheme of human origins, but long enough that the rockshelter was home to several different groups with different genetic connections.
In 2020, Mark Lipson and collaborators reported on DNA from Shum Laka. In some of the earliest burials were people with genetic ties to living Aka people from slightly further east. Putting these genomes into a tree with other ancient and modern African populations, Lipson and coworkers found that a single ghost population was not enough to explain the genetic data. They proposed two: One “ghost archaic” similar to that proposed by Durvasula and Sankararaman, and one “ghost modern” that had begun to differentiate some 250,000 years ago or so.
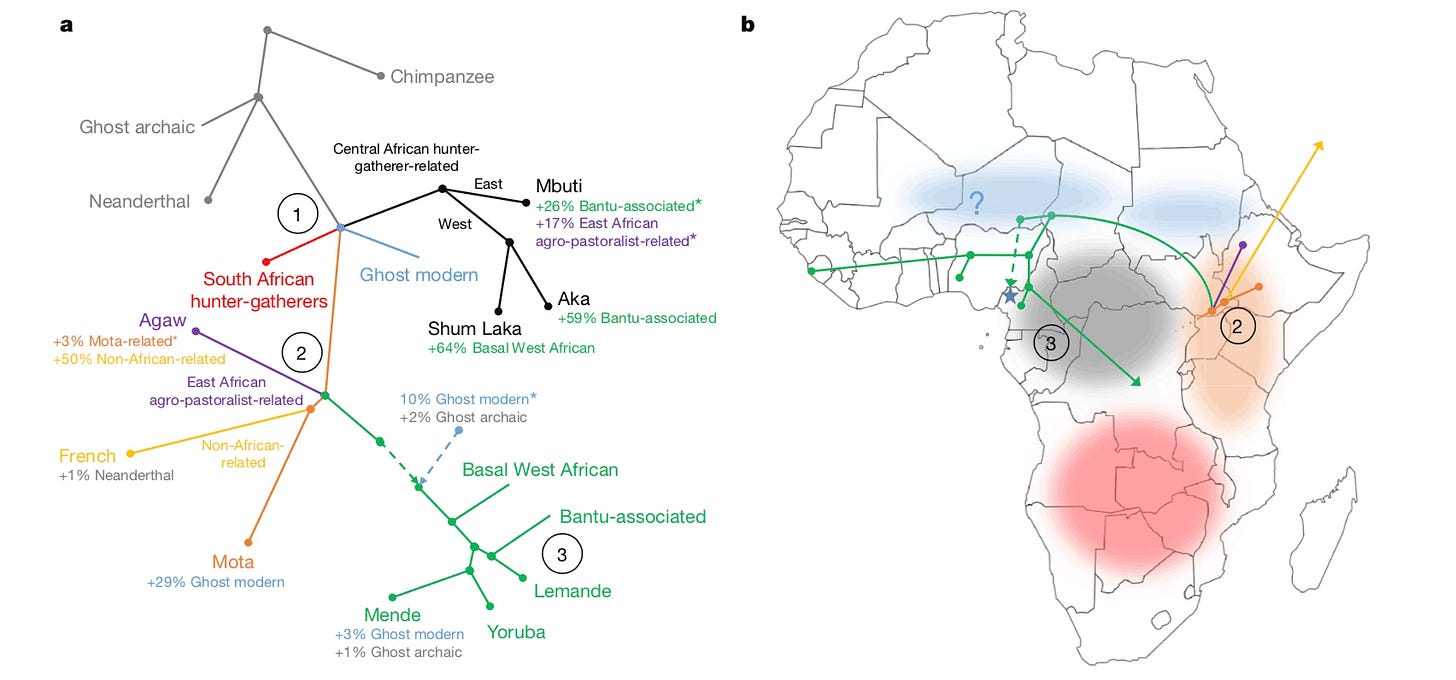
Ancestral North Africans
Earlier this year, researchers from Johannes Krause’s research group led by Nada Salem reported on DNA that they pulled from skeletons that were excavated from Takarkori rock shelter in the Libyan desert. These people were part of an early herding society around 7000 years ago. The area was then a well-watered savanna with rivers and areas of woodland, very different from the arid desert environment of today. The DNA of these people suggests that more than 90% of their ancestry came from a deep North African lineage that no longer exists.
Geneticists will probably call this ancestral group the Takarkori population from now on. Or maybe they’ll settle instead on Ancestral North Africans. With such a high proportion of DNA in these skeletons, I wouldn’t say their previously-unknown ancestors are a ghost population anymore.
But they were one until now. Fifteen thousand years ago, ancient foraging peoples lived and buried their dead in Grotte des Pigeons, near Taforalt, Morocco. Skeletons were unearthed there by archaeologists working from the 1940s intermittently up to the 1970s. In 2018, Krause’s group led by Marieke van de Loosdrecht reported on DNA from seven of those skeletons. They found that a fraction of the DNA of these ancient people came from an unknown source population, a ghost. The new work from Takarkori has revealed the ghost: the same ancestral North African lineage is ancestor to both of these groups.
The antiquity of this Takarkori branch is impressive: dating back as far as the initial founding population of non-African peoples around 50,000 years ago. I expect that working out the history of this branch may be a key to the history of that founding population before it began the great dispersal across Eurasia and toward the rest of the world.
Population Y
One of the best-known ghost populations pertains to the ancestry of Indigenous peoples of the Amazon Basin. In 2015, Pontus Skoglund and collaborators found that some groups from this region have a slight but noticeable genetic similarity with living peoples of Australia, Papua, and the Andaman Islands. In this, the Amazonian peoples are outliers. Other groups sampled across the Americas do not show this same signature.
The team posited that the early founding populations of the Americas had at least two different histories. One of those histories had been connected with early founders of populations now living in the Andaman Islands and Sahul. The researchers began to call this founding group Population Y, from a word meaning “ancestor” in the Tupi language, ypykuéra.
Some anthropologists liked the Population Y idea, especially those who perceived possible southeast Asian similarities in a handful of early cranial and skeletal remains from the Americas. Ancient DNA from South America is still scarce, but genome data from some skeletons are consistent Population Y ancestry, including several individuals from Lagoa Santa, Brazil, dating to around 10,400 years ago, as well as several skeletons from coastal Brazil from between 3200 to 2400 years ago. In the 2018 paper that reported the DNA from Lagoa Santa, Victor Moreno-Mayar and coworkers suggested that 3% of the ancestry of these groups came from a “Australasian-like” population related to Andaman Island peoples.
Still, there are researchers who are skeptical of the Population Y scenario. In their view, the appearance of a small fraction of Andaman-like ancestry might be an illusion. This point of view comes down to the methods that geneticists use to infer ancient mixture. A close look at those methods shows there is plenty of room for uncertainty.
How real were the ghosts?
As I mentioned at the top, a ghost population is no more nor less than a mathematical construct. When geneticists plug a set of DNA samples into a model to understand their connections, sometimes the model spits out an unknown component, a ghost.
“All models are wrong, but some models are useful.” What George Box wrote about statistical models holds true in this case.
Some models are easy to build and test. Easiest are models with populations that stay isolated after they split, except for the rare times of mixing between them. These models operate a bit like populations that lived on islands, and geneticists call them “island models”. When such models spit out a lineage, it looks like a ghost.
But humans across Africa and Eurasia during the Pleistocene did not live on a handful of islands. If their genomes have few signs of mixing over time, it’s not because none of them ever met. More likely, their long-term evolution was driven by growth in some regions much more than others. Places where different groups met were not often centers of any population’s growth.
Two groups on two islands, or different rates of growth and limits on mixture in different parts of a single population’s range. These are factors that geneticists call population structure.
For many years, some geneticists were skeptical of the idea that divergent haplotypes in living people might be a legacy of Neanderthal ancestors. They noted that certain kinds of population structure in ancient Africans might also result in such haplotypes. It took an array of DNA data from Neanderthals and better knowledge of haplotype variation in living humans to accept Neanderthal ancestors.
Ancestral population structure in Africa may be an alternative to the “ghost archaics”. Aaron Ragsdale and coworkers in 2023 took a critical look at the ghost population scenario for African ancestral groups. The found that a population structure model, with continuous mixing between populations, can also explain data from living African peoples. A similar argument was suggested by Tiago Ferraz and collaborators for the Population Y hypothesis in South America: Population structure within a single founder group of populations in the Americas, not a second founder population.
Still, I’ve become a believer in ghosts. We know that every model is wrong in some ways. The ghost model is often a useful one. The point of talking about ghosts is to remind us of the past realities that we cannot see with today’s data.
And to be honest, unknown population structure looks an awful lot like a ghost. If African groups began to differentiate more than a million years ago, and today’s African people get 95% of their ancestry from one part of that early population and 5% from others, that sounds a lot like a ghost population. If early modern humans diversified rapidly across much of Africa when they emerged 250,000 years ago, but the groups that came to occupy one area are only visible as a small fraction of the genomes of today’s people, that sounds a lot like a ghost population.
So I’ve come to like the term “ghost population” quite a lot. There’s only one thing that I try to remind people. These ghosts are not dead. Theirs is a legacy of genetic persistence, of mixing into groups where their descendants survived. Ghost populations are ancestors we haven’t recognized yet.
Notes: I’ve written a good amount about ghost populations over the last couple of years. The post on ghost populations in the evolutionary history of living great apes helps give some broader context to the conversation in human ancestry. Two years ago I wrote about the work of Aaron Ragsdale and colleagues, which challenges the idea of ghost populations in the Middle and Late Pleistocene of Africa: “Ghostbusters of human origins”.
Sometime I’ll expand more on the story of my own work uncovering Neanderthal introgression in today’s populations. The case of MCPH, which seemed like such a promising case study, turns out to be not so simple: No one has yet found the divergent haplotype in an ancient genome. Probably not a ghost; there’s a good possibility that this haplotype was conserved over time in humans by balancing selection.
References
Durvasula, A., & Sankararaman, S. (2020). Recovering signals of ghost archaic introgression in African populations. Science Advances, 6(7), eaax5097. https://doi.org/10.1126/sciadv.aax5097
Ferraz, T., Suarez Villagran, X., Nägele, K., Radzevičiūtė, R., Barbosa Lemes, R., Salazar-García, D. C., Wesolowski, V., Lopes Alves, M., Bastos, M., Rapp Py-Daniel, A., Pinto Lima, H., Mendes Cardoso, J., Estevam, R., Liryo, A., Guimarães, G. M., Figuti, L., Eggers, S., Plens, C. R., Azevedo Erler, D. M., … Posth, C. (2023). Genomic history of coastal societies from eastern South America. Nature Ecology & Evolution, 7(8), 1315–1330. https://doi.org/10.1038/s41559-023-02114-9
Lachance, J., Vernot, B., Elbers, C. C., Ferwerda, B., Froment, A., Bodo, J.-M., Lema, G., Fu, W., Nyambo, T. B., Rebbeck, T. R., Zhang, K., Akey, J. M., & Tishkoff, S. A. (2012). Evolutionary History and Adaptation from High-Coverage Whole-Genome Sequences of Diverse African Hunter-Gatherers. Cell, 150(3), 457–469. https://doi.org/10.1016/j.cell.2012.07.009
Lipson, M., Ribot, I., Mallick, S., Rohland, N., Olalde, I., Adamski, N., Broomandkhoshbacht, N., Lawson, A. M., López, S., Oppenheimer, J., Stewardson, K., Asombang, R. N., Bocherens, H., Bradman, N., Culleton, B. J., Cornelissen, E., Crevecoeur, I., de Maret, P., Fomine, F. L. M., … Reich, D. (2020). Ancient West African foragers in the context of African population history. Nature, 577(7792), Article 7792. https://doi.org/10.1038/s41586-020-1929-1
Moreno-Mayar, J. V., Vinner, L., de Barros Damgaard, P., de la Fuente, C., Chan, J., Spence, J. P., Allentoft, M. E., Vimala, T., Racimo, F., Pinotti, T., Rasmussen, S., Margaryan, A., Iraeta Orbegozo, M., Mylopotamitaki, D., Wooller, M., Bataille, C., Becerra-Valdivia, L., Chivall, D., Comeskey, D., … Willerslev, E. (2018). Early human dispersals within the Americas. Science, 362(6419), eaav2621. https://doi.org/10.1126/science.aav2621
Patterson, N., Moorjani, P., Luo, Y., Mallick, S., Rohland, N., Zhan, Y., Genschoreck, T., Webster, T., & Reich, D. (2012). Ancient Admixture in Human History. Genetics, 192(3), 1065–1093. https://doi.org/10.1534/genetics.112.145037
Prüfer, K., Racimo, F., Patterson, N., Jay, F., Sankararaman, S., Sawyer, S., Heinze, A., Renaud, G., Sudmant, P. H., de Filippo, C., Li, H., Mallick, S., Dannemann, M., Fu, Q., Kircher, M., Kuhlwilm, M., Lachmann, M., Meyer, M., Ongyerth, M., … Pääbo, S. (2014). The complete genome sequence of a Neanderthal from the Altai Mountains. Nature, 505(7481), 43–49. https://doi.org/10.1038/nature12886
Rogers, A. R., Harris, N. S., & Achenbach, A. A. (2020). Neanderthal-Denisovan ancestors interbred with a distantly related hominin. Science Advances, 6(8), eaay5483. https://doi.org/10.1126/sciadv.aay5483
Salem, N., van de Loosdrecht, M. S., Sümer, A. P., Vai, S., Hübner, A., Peter, B., Bianco, R. A., Lari, M., Modi, A., Al-Faloos, M. F. M., Turjman, M., Bouzouggar, A., Tafuri, M. A., Manzi, G., Rotunno, R., Prüfer, K., Ringbauer, H., Caramelli, D., di Lernia, S., & Krause, J. (2025). Ancient DNA from the Green Sahara reveals ancestral North African lineage. Nature, 641(8061), 144–150. https://doi.org/10.1038/s41586-025-08793-7
Skoglund, P., Mallick, S., Bortolini, M. C., Chennagiri, N., Hünemeier, T., Petzl-Erler, M. L., Salzano, F. M., Patterson, N., & Reich, D. (2015). Genetic evidence for two founding populations of the Americas. Nature, 525(7567), 104–108. https://doi.org/10.1038/nature14895
van de Loosdrecht, M., Bouzouggar, A., Humphrey, L., Posth, C., Barton, N., Aximu-Petri, A., Nickel, B., Nagel, S., Talbi, E. H., El Hajraoui, M. A., Amzazi, S., Hublin, J.-J., Pääbo, S., Schiffels, S., Meyer, M., Haak, W., Jeong, C., & Krause, J. (2018). Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science, 360(6388), 548–552. https://doi.org/10.1126/science.aar8380



Maybe we will find some of these mysterious ancestors in the future. Based on this article, I have decided to believe in ghosts too.
It seems to me that a few decades ago, anthropologists believed that Homo sapiens was a descendant of Neanderthals.
This hypothesis was abandoned, but I haven't been able to find any papers that clearly show that Homo sapiens was not a descendant of Neanderthals.
Yet, during the supposed period of their cohabitation, skeletons and DNA were found with traits shared by each species. Wouldn't this show that Neanderthals were evolving into Homo sapiens?
For how can we explain why Neanderthals, who lived for several hundred thousand years on the Eurasian continent, would have disappeared in less than 10,000 years?
On the other hand, I can't understand what it clearly means that we have between 1 and 4% of the Neanderthal genome in Europe.