The humanity of a new Denisovan
Identification of the Harbin skull by mtDNA and proteins and what it means to be Homo sapiens.
Last week a team led by Qiaomei Fu revealed a partial mitochondrial DNA sequence from the Harbin skull. In a second paper, Fu and her team also described protein evidence from the skull. Together, the mtDNA and protein evidence embed the Harbin individual within a broader lineage that we know as “Denisovans”.
I’m very excited about these new results. They clarify some things about the Denisovans and enable some new predictions about this group and the broader fossil record of the region. At the same time, the findings are narrow: Mitochondrial DNA represents only one of the Harbin individual’s lines of ancestry and protein data have only a limited ability to link fossils to each other. I’ll share some of my thinking, especially about how we talk about these ancient groups.
Let me start with the question the New York Times asked me: What do we call them? I call them Homo sapiens. Just like all living people, the Denisovans are part of our species. The history of repeated interbreeding among the Denisovans, Neanderthals, and their contemporaries in Africa shows that while geography may have made them look a little different, their shared biology went deep enough to bring them together into our genetic heritage.
Harbin and the Denisovans
This ancient skull from northeastern China was first described in 2021 by Xijun Ni and collaborators, and given the name Homo longi by Qiang Ji and coworkers, including Ni. The fossil had a very strange history. It was first brought to scientific attention in 2018 by an anonymous person with a strange tale. While the original research paper in 2021 told the story as fact, Dyani Lewis reported in Nature this week that it may be fiction.
“The man — who Ji suspects discovered the artefact himself but failed to report it to authorities — claimed that his grandfather unearthed the fossil in 1933 during bridge-construction work over Long Jiang (which means dragon river), and buried it in an abandoned well, where it remained until a deathbed confession.”—Dyani Lewis
Due in part to the skull’s dodgy provenience, its geological age is not precisely known. The limited evidence suggests that the individual lived at least 146,000 years ago.

Denisovans got their name from Denisova Cave, Russia, where DNA evidence from fossil fragments has been extraordinarily well preserved. One individual, Denisova 3, has a high-coverage genome. A small number of fossils from other sites have been linked to the Denisovans, including a partial jawbone and rib from Baishiya Karst Cave in western China and a partial jaw dredged from the sea bottom near the Penghu Islands.
Scientists began to understand that the Denisovan population might be a widespread genetic phenomenon almost as soon as the Denisova 3 genome was sequenced. Living people in many parts of the world have segments of their genomes that are closer to this ancient individual than to other living people or to Neanderthals. The presence of this DNA legacy in Papua, Australia, the Philippines, and more broadly across mainland East and Southeast Asia and Oceania makes it clear that modern people derive a small part of their ancestry from this group.
Until now, no fossil with a face or cranium had been attributed to this group. Fu and collaborators found that the Harbin individual fits within the mtDNA variation of the fossil fragments from Denisova Cave. Harbin is closer in relationship to earlier Denisovans than with the two latest fossil individuals, Denisova 3 and Denisova 4.
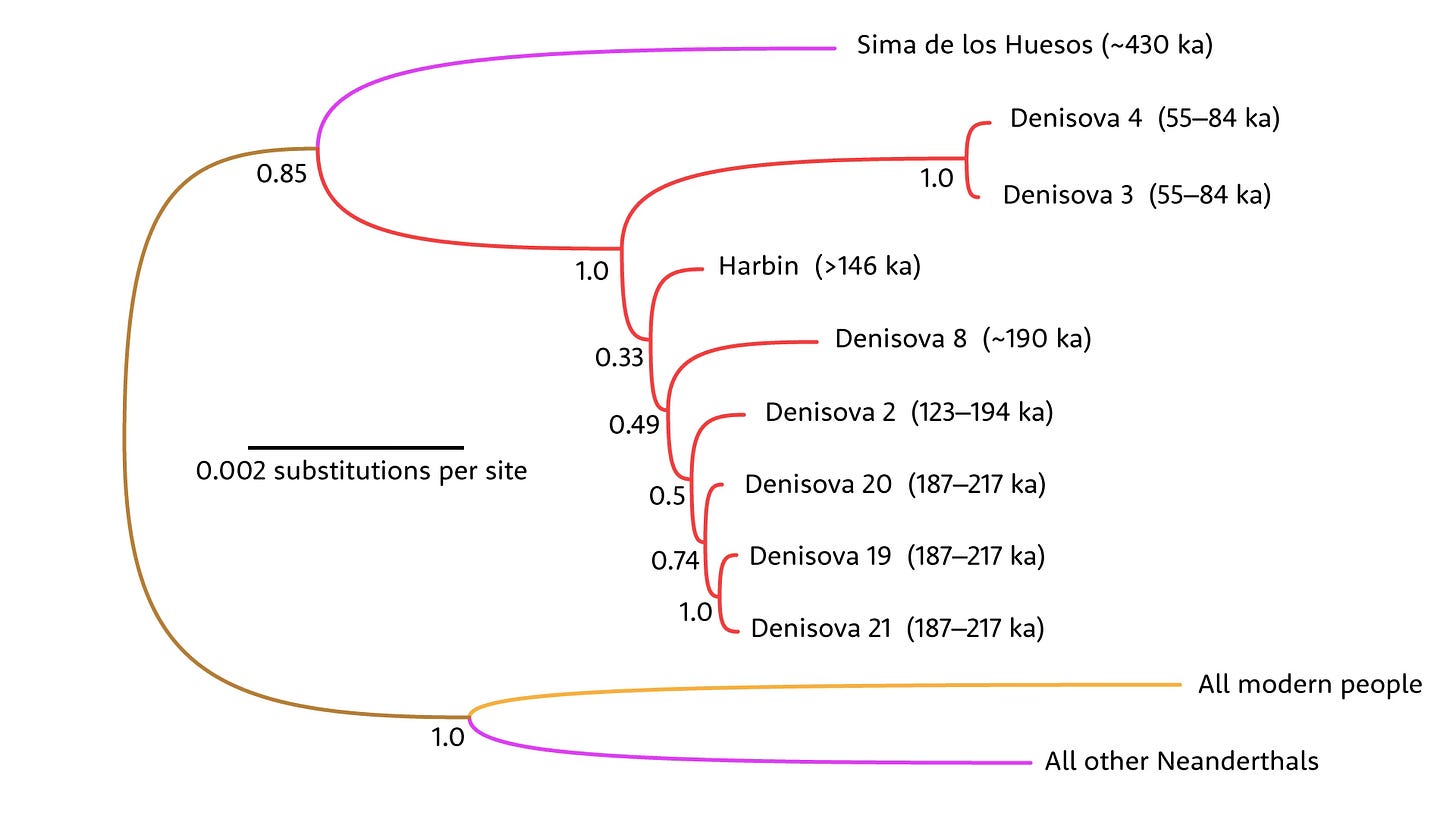
Population structure analyses that have brought light to Neanderthal deep history are more challenging for Denisovans. The lack of sites with DNA other than Denisova and Baishiya Karst Cave means geneticists can’t look for major transitions across space, and the single high-coverage genome of Denisova 3 enables only limited probing into ancient interactions.
Still, there are signs of complexity. The Denisova Cave sediment DNA sequence shows that a series of alternating Neanderthal and Denisovan populations entered the site over hundreds of thousands of years. Mitochondrial DNA has been taken from many hominin bone fragments from Denisova Cave. A few of those are Neanderthals, like the Denisova 5 “Altai” Neanderthal. Others from the cave are Denisovans, and these fragments have a pattern in which later Denisovans had limited mtDNA variation compared to earlier ones.
That suggests some kind of population transition happened, with the later samples representing a different source population than earlier ones. This transition may have occurred across a broader region: Sediment DNA samples from Baishiya Karst Cave, all between around 100,000 and 32,000 years old, have mtDNA sequences closer to the later Denisova fossils.
Few are really saying this, but if the Harbin skull belongs to the broader Denisovan population, it’s very likely that all or nearly all of the fossils from China during the period from 350,000 to 130,000 years ago also belong to this group. Although each fossil has its own story, and some researchers have emphasized differences that might be deep population history within this group, I think their diversity is more or less what we might expect from populations that ebbed, flowed, and mixed in the way DNA suggests.
Mixture in the past
The broad story of ancient DNA over the last 15 years has been the extent of mixture among these ancient groups. Several low-coverage genomes from Denisovan Cave and Chagyrskaya Cave document the repeated intermixture between Denisovan and Neanderthal populations in this region of central Asia. Almost every genome preserves this legacy of mixed ancestry, ranging from a few generations to tens of thousands of years before. The single most evocative evidence comes from the Denisova 11 individual, born from a Neanderthal mother and Denisovan father.
The evidence from the Harbin skull does not rule out the idea that this individual had an ancestry with mixture among varied groups. That’s a possibility that interests me because of what we know about Neanderthal ancestry and mixture in western and central Eurasia.
At least one major influx of genes from African people—or more precisely, people sharing DNA similarity with the African ancestors of today’s humans—spread across Neanderthal groups sometime before 250,000 years ago. The result: Later Neanderthals trace every bit of their mtDNA and Y chromosome variation, and an estimated 6% of their whole genomes, to those African groups.
So far nobody has shown Denisovan gene segments within the few Neanderthal genomes that have been sampled far to the west. With as much mixture as there was in the Denisova Cave region, one possibility is that there was more migration toward the Altai than away from it—known as a population sink. Another possibility is that the channels of gene flow went toward the south, linking up with Neanderthal populations in present-day Afghanistan, Iran, and Iraq. Until DNA evidence comes from such areas we may not know.
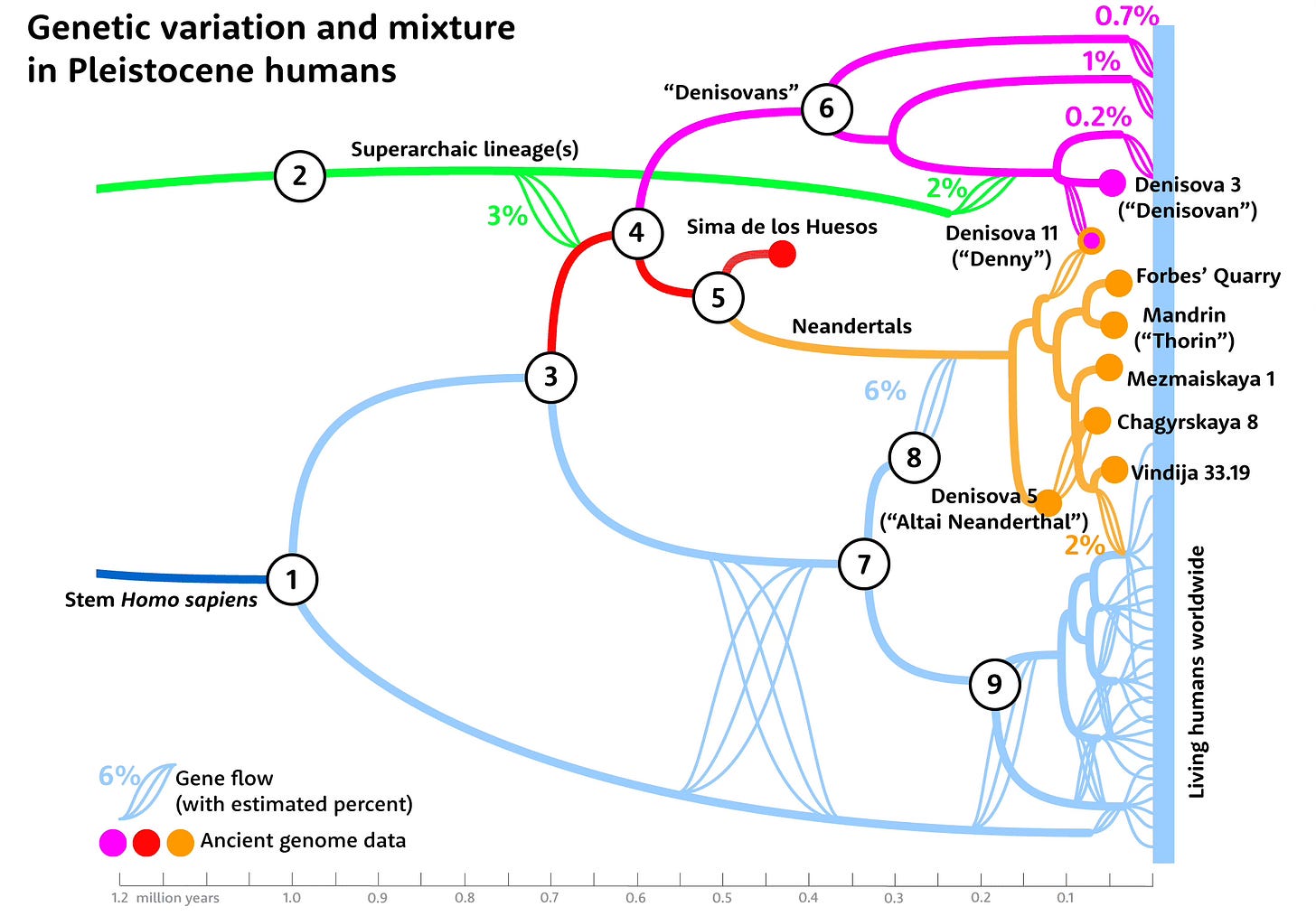
Could Neanderthal genes have entered China? Some researchers have suggested that Neanderthals were part of the ancestral makeup of archaic humans in the area, in part because they have noted some individuals with features recognized in Neanderthals. No DNA evidence has emerged yet—but then, Harbin is the first fossil older than the modern human Tianyuan skeleton to yield any of its DNA.
On the subject of mixture, there is also the idea of a “superarchaic ghost” contribution to the Denisova 3 genome. Based on sequence divergence of introgressed genome segments, the Denisovans might have encountered, coexisted, and mixed with late-surviving groups of Homo erectus. A recent preprint by Xiaobo Feng and coworkers suggested something like this scenario, as it linked the anatomy of the one-million-year-old Yunxian 2 Homo erectus cranium to the much later Harbin skull. Maybe the Dragon people had an autochthonous tendril stretching beyond the Denisovan branch.
The breadth of Homo sapiens
Nobody reading my work is a stranger to the idea that today’s people have a mixed heritage going back through the Pleistocene. I’ve been writing a lot about work showing the deep population structure evident by studying today’s peoples. Among the traditional peoples of many parts of the world, the most evident contributions of deep structure come from Neanderthals or Denisovans. The deep African components of structure were more homogenized by the process of founder effects that led to African dispersal into Eurasia and beyond.
Within Africa today the signs of deep structure from ancestors from humankind’s original home survive in forms that geneticists are beginning to understand. Today’s peoples of Africa did not come recently from a single small group, but instead emerged from the repeated mixture of ancient populations.
The pattern of this mixture across Pleistocene Africa is not yet clear. Some geneticists have worked with models of “ghost populations” that were isolated for hundreds of thousands of years before ultimately mixing. Others—like my colleague Aaron Ragsdale—have explored less extreme models with more frequent mixture that fit the genetic data equally well.
What all the models share is that the initiation of this pattern of deep human diversity goes back roughly a million years. That’s long before the branching of Denisovan and Neanderthal ancestors from Africa, around 700,000 years ago. Ghost or not, from that time on humanity was a braided stream.
When I look at my own genome, there are segments on my own chromosomes where my two copies are very nearly identical, differing by only one or two base pairs out of ten thousand. For those segments, my two genealogical lineages—from my mother and my father—must trace back to an ancestor within the last thousand or so generations of time. But for most segments of DNA, my two copies are more different, nearly one base pair out of a thousand will mismatch. Each of those genomic swatches traces to a different common ancestor, most of which lived between roughly 350,000 and 1.2 million years ago.
This is the origin of present-day differences. Not the differences between groups dispersed around the world, which are tiny in comparison. The differences between the genomic halves within every one of us. The mainstream variation of humanity today emerged during the early differentiation of deep African groups, and the early branching of the Neanderthals and Denisovans. What is different is shared. This is why I call these ancestors our own species, Homo sapiens.
What’s in a name
I have close colleagues who see morphology as the guide to understanding ancient species. If derived traits reliably separate ancient fossil groups, that is the criterion for recognizing a species under the phylogenetic species concept. It’s in our introductory courses. The sorting of fossil groups into named classification has been a big aim of paleontology, and paleoanthropologists are among the most eager namers of all.
For these friends, the Harbin skull has a special kind of importance. It may help connect dozens of observable cranial traits to the genomic variants carried in the Denisova 3 genome. It also may help to confirm that work by David Gokhman and coworkers on the methylation of ancient genomes might really provide some guide to the morphology of ancient hominins. The attribution of the Harbin individual to Homo longi is for them a starting point to classification.
Others of this bent may find different names a better basis for classification. A more expansive Homo neanderthalensis, encompassing not just the classic Neanderthals but variation extending to Sima de los Huesos and Harbin. Or maybe Homo soloensis, attributing the 110,000-year-old fossil group from Ngandong, Indonesia, to the Denisovan branch. Even the Penghu jaw, aligned this year with Denisovans by its proteomic profile, has a name: Homo tsaichangensis, given by Mark McMenamin in 2015, in a publication most classification aficionados reject as insufficiently reviewed.
I’ll be honest, I think that such names obscure the vital point. The course of our evolution was shaped by the mixing of these populations, the adaptations to varied environments where they lived, and their sharing with each other. That’s what species should mean in our evolutionary history, and that’s why I recognize their shared nature within our species.
These ancient humans from different backgrounds didn’t have a chance to meet each other as often as we do today. They would have had very different cultures, whatever means of speaking they had would have been different. But they learned from each other and mated when they met. They and we are part of a single extended genealogy. That network may sometimes have been stretched but was not broken.
Notes: When I raised this idea of interbreeding as a basis for naming Homo sapiens with a colleague, they asked why I would consider Pleistocene fossils like Homo naledi or Homo floresiensis to be something different. The answer to that is its own post, but in brief I think it is very likely that these hominins did not interbreed with humans when they met. The fact that H. naledi in particular may have been quite behaviorally sophisticated—sometimes in ways that resemble modern humans or Neanderthals, and sometimes in its own ways that are quite different—is important to view in this light. I think they were not us. Maybe DNA will someday prove me wrong about that.
References
Bae, C. J., Liu, W., Wu, X., Zhang, Y., & Ni, X. (2023). “Dragon man” prompts rethinking of Middle Pleistocene hominin systematics in Asia. The Innovation, 4(6), 100527. https://doi.org/10.1016/j.xinn.2023.100527
Chen, F., Welker, F., Shen, C.-C., Bailey, S. E., Bergmann, I., Davis, S., Xia, H., Wang, H., Fischer, R., Freidline, S. E., Yu, T.-L., Skinner, M. M., Stelzer, S., Dong, G., Fu, Q., Dong, G., Wang, J., Zhang, D., & Hublin, J.-J. (2019). A late Middle Pleistocene Denisovan mandible from the Tibetan Plateau. Nature, 569(7756), Article 7756. https://doi.org/10.1038/s41586-019-1139-x
Feng, X., Lu, D., Gao, F., Fang, Q., Feng, Y., Huang, X., Tan, C., Zhou, H., Li, Q., Zhang, C., Stringer, C., & Ni, X. (2024). The phylogenetic position of the Yunxian cranium elucidates the origin of Dragon Man and the Denisovans(p. 2024.05.16.594603). bioRxiv. https://doi.org/10.1101/2024.05.16.594603
Fu, Q., Bai, F., Rao, H., Chen, S., Ji, Y., Liu, A., Bennett, E. A., Liu, F., & Ji, Q. (2025a). The proteome of the late Middle Pleistocene Harbin individual. Science, 0(0), eadu9677. https://doi.org/10.1126/science.adu9677
Fu, Q., Cao, P., Dai, Q., Bennett, E. A., Feng, X., Yang, M. A., Ping, W., Pääbo, S., & Ji, Q. (2025b). Denisovan mitochondrial DNA from dental calculus of the >146,000-year-old Harbin cranium. Cell, 0(0). https://doi.org/10.1016/j.cell.2025.05.040
Ji, Q., Wu, W., Ji, Y., Li, Q., & Ni, X. (2021). Late Middle Pleistocene Harbin cranium represents a new Homo species. The Innovation, 2(3). https://doi.org/10.1016/j.xinn.2021.100132
Lewis, D. (2025). First ever skull from ‘Denisovan’ reveals what ancient people looked like. Nature. https://doi.org/10.1038/d41586-025-01899-y
Li, Z.-Y., Wu, X.-J., Zhou, L.-P., Liu, W., Gao, X., Nian, X.-M., & Trinkaus, E. (2017). Late Pleistocene archaic human crania from Xuchang, China. Science, 355(6328), 969–972. https://doi.org/10.1126/science.aal2482
Mafessoni, F., Grote, S., de Filippo, C., Slon, V., Kolobova, K. A., Viola, B., Markin, S. V., Chintalapati, M., Peyrégne, S., Skov, L., Skoglund, P., Krivoshapkin, A. I., Derevianko, A. P., Meyer, M., Kelso, J., Peter, B., Prüfer, K., & Pääbo, S. (2020). A high-coverage Neandertal genome from Chagyrskaya Cave. Proceedings of the National Academy of Sciences, 117(26), 15132–15136. https://doi.org/10.1073/pnas.2004944117
Massilani, D., Skov, L., Hajdinjak, M., Gunchinsuren, B., Tseveendorj, D., Yi, S., Lee, J., Nagel, S., Nickel, B., Devièse, T., Higham, T., Meyer, M., Kelso, J., Peter, B. M., & Pääbo, S. (2020). Denisovan ancestry and population history of early East Asians. Science, 370(6516), 579–583. https://doi.org/10.1126/science.abc1166
McMenamin, M. (2015). Homo tsaichangensis and Gigantopithecus. Meanma Press.
Mishol, N., Herzlinger, G., Rak, Y., Smilanksy, U., Carmel, L., & Gokhman, D. (2024). Candidate Denisovan fossils identified through gene regulatory phenotyping (p. 2024.04.18.590145). bioRxiv. https://doi.org/10.1101/2024.04.18.590145
Ni, X., Ji, Q., Wu, W., Shao, Q., Ji, Y., Zhang, C., Liang, L., Ge, J., Guo, Z., Li, J., Li, Q., Grün, R., & Stringer, C. (2021). Massive cranium from Harbin in northeastern China establishes a new Middle Pleistocene human lineage. The Innovation, 2(3). https://doi.org/10.1016/j.xinn.2021.100130
Ragsdale, A. P., Weaver, T. D., Atkinson, E. G., Hoal, E. G., Möller, M., Henn, B. M., & Gravel, S. (2023). A weakly structured stem for human origins in Africa. Nature, 1–9. https://doi.org/10.1038/s41586-023-06055-y
Skov, L., Peyrégne, S., Popli, D., Iasi, L. N. M., Devièse, T., Slon, V., Zavala, E. I., Hajdinjak, M., Sümer, A. P., Grote, S., Bossoms Mesa, A., López Herráez, D., Nickel, B., Nagel, S., Richter, J., Essel, E., Gansauge, M., Schmidt, A., Korlević, P., … Peter, B. M. (2022). Genetic insights into the social organization of Neanderthals. Nature, 610(7932), 519–525. https://doi.org/10.1038/s41586-022-05283-y
Wu, X., Pei, S.-W., Cai, Y.-J., Tong, H.-W., Li, Q., Dong, Z., Sheng, J.-C., Jin, Z.-T., Ma, D.-D., Xing, S., Li, X.-L., Cheng, X., Cheng, H., de la Torre, I., Edwards, R. L., Gong, X.-C., An, Z.-S., Trinkaus, E., & Liu, W. (2019). Archaic human remains from Hualongdong, China, and Middle Pleistocene human continuity and variation. Proceedings of the National Academy of Sciences, 116(20), 9820–9824. https://doi.org/10.1073/pnas.1902396116
Wu, Xiujie. (2024). Research progress on human fossils from the Xujiayao site in late Middle Pleistocene. Acta Anthropologica Sinica, 43(01), 5. https://doi.org/10.16359/j.1000-3193/AAS.2023.0044
Wu, Xiujie & Bae, Christopher J. (2024). Xujiayao Homo: A New Form of Large Brained Hominin in Eastern Asia. PaleoAnthropology, 2024. https://paleoanthropology.org/ojs/index.php/paleo/libraryFiles/downloadPublic/18
Zhang, D., Xia, H., Chen, F., Li, B., Slon, V., Cheng, T., Yang, R., Jacobs, Z., Dai, Q., Massilani, D., Shen, X., Wang, J., Feng, X., Cao, P., Yang, M. A., Yao, J., Yang, J., Madsen, D. B., Han, Y., … Fu, Q. (2020). Denisovan DNA in Late Pleistocene sediments from Baishiya Karst Cave on the Tibetan Plateau. Science, 370(6516), 584–587. https://doi.org/10.1126/science.abb6320
Cite as: https://doi.org/TK • PDF



(Continued). These dates (~1.5 mY and ~300 kY ago) remarkably coincide with big steps in stone age technology. The 1st one marks the time the Acheulean (or “mode 2”) technology appeared in Eurasia – in Middle East and in India – while in Africa it began earlier (~1.7 mY ago). So, this might be the sign of exit from Africa of carriers of this technology (and emergence of “80% branch” or “stem 1” of previous comment). This might be the last true “exit from Africa” of HS ancestors.
The second date is approximate time of appearance of Levallois technology and the beginning of the MP. It likely appeared in W. Eurasia first and led to population growth and spread into Africa, where it merged with the “20% branch”. So, after ~300 kY ago there appeared a unified Eurasian-African population of early modern humans. It is interesting that there are parallel fossil African-Eurasian pairs across huge distances that have similar looks and archaic/modern feature combinations: Jebel Irhoud and Hualongdong (~300 kY) or Eli Spring (Kenya) and Dali (China) ~250 kY. Probably this unified population were some variants of H. heidelbergensis.
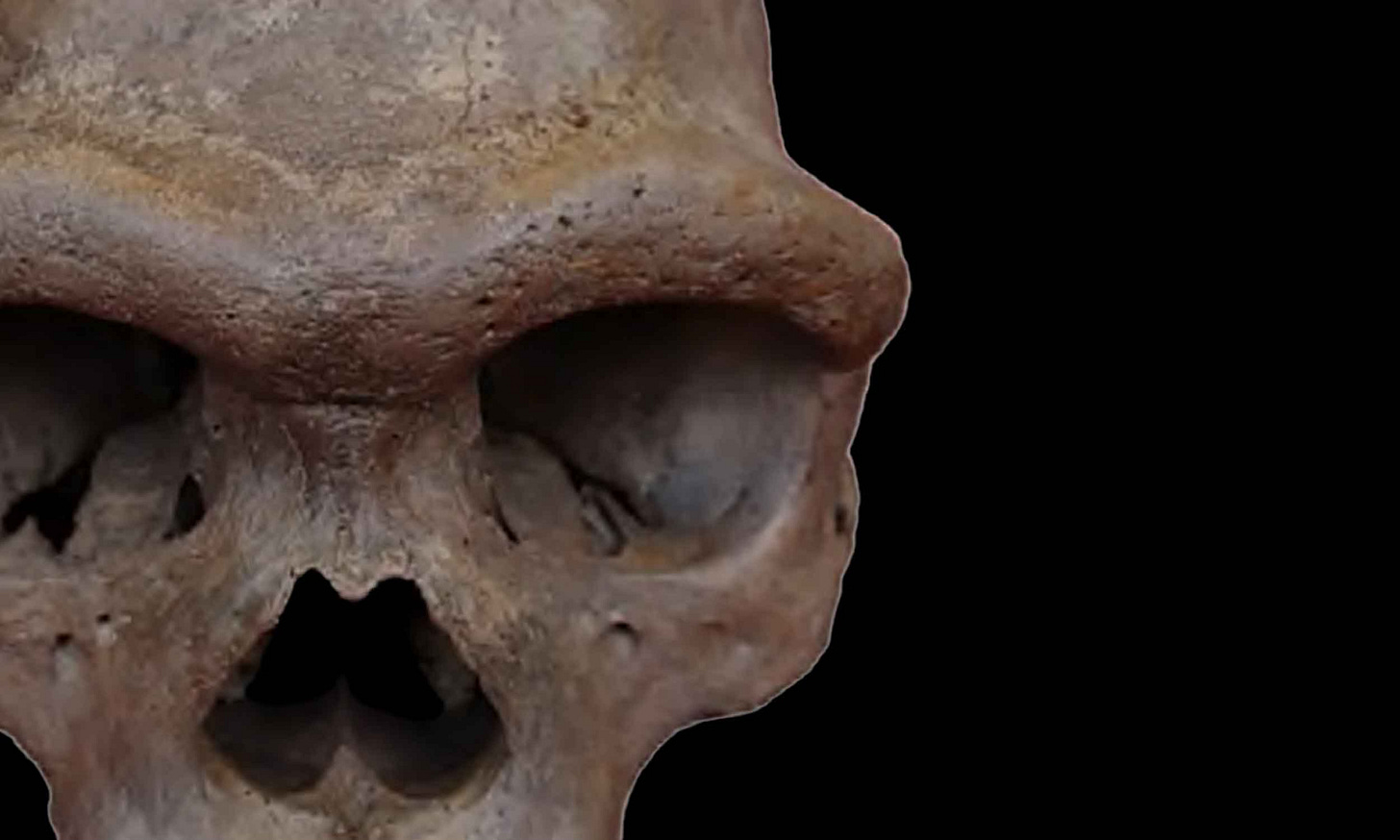
It is very exciting to put some “face” on abstract genomic data of “Denisovans”.
I think it is a true revolution in paleoanthropology in the last 15 years – the knowledge that there were no firm genetic barriers between any homo branches, even after >1mY separation. As recently as 2009 the dogma was that HS, Neanderthals and erectuses were completely separate species.
One interesting development in the last couple of years has been the “deep structure” present in modern HS, and two much-quoted papers “A structured coalescent model reveals deep ancestral structure shared by all modern humans” (Cousins et al, 2025) and a “two stems” model (Ragsdale et al, 2023), as well as many works on “ghost populations” point to a similar thing. There was a very ancient genetic separation between ancestral HS branches (1.5 mY in Cousins et all, 1.0-1.2 mY “stem 1 - stem 2 split” in Ragsdale et al, (also the node (1) in the picture in this post), and much later (~300 kY ago) the merger of these branches (or merger of “stem 2” with “stem 1E” and “stem 1s” in Ragsdale et al).
If these models are relatively correct, I don’t believe a genetic barrier could be maintained for ~1mY within a single continent. These two branches most likely existed across the narrowest divide – Africa – SW Asia corridor. Then the “stem 1” and the Cousins et al 80% branch (that included Neanderthals and Denisovans) was likely in Eurasia, and the “stem 2” or 20% branch – in Africa. From that it would follow that most of the modern HS ancestry is in fact the Eurasian one.
The “2 stems” model of Ragsdale et al is unnecessarily convoluted, I believe, mainly to avoid the notion of “African superarchaic” population (likely for ideological reason), which would make the model simpler, instead of it being a weird “subway map”.